Genie 2: Transforming Protein Design with Advanced Multi-Motif Scaffolding and Enhanced Structural Diversity
Protein design is a rapidly advancing field leveraging computational models to create proteins with novel structures and functions. This technology has significant applications in therapeutics and industrial processes, revolutionizing how proteins are engineered for specific tasks. Researchers in this field aim to develop methods that accurately predict and generate protein structures that perform desired functions efficiently. The complexity of protein folding and interaction dynamics presents a significant challenge, making it crucial to innovate in this space.
Designing proteins with precise structural and functional properties remains challenging. The primary objective is to create proteins that perform specific functions, such as enzyme catalysis or molecular recognition, essential in various biological and industrial applications. The intricate nature of protein structures, composed of amino acids folding into three-dimensional shapes, necessitates advanced computational tools to accurately predict and design these configurations.
Current methods in protein design include sequence-based and structure-based approaches. Sequence-based models, such as EvoDiff, predict amino acid sequences that fold into functional proteins, while structure-based models like ProteinMPNN propose plausible sequences for given structures. However, these methods often need help designing proteins involving multiple interaction sites. For example, RFDiffusion integrates sequence information as a condition of a structure-based diffusion process, and FrameFlow combines a structural flow with a sequence flow. Designing proteins with multiple independent motifs remains a significant hurdle despite these advancements.
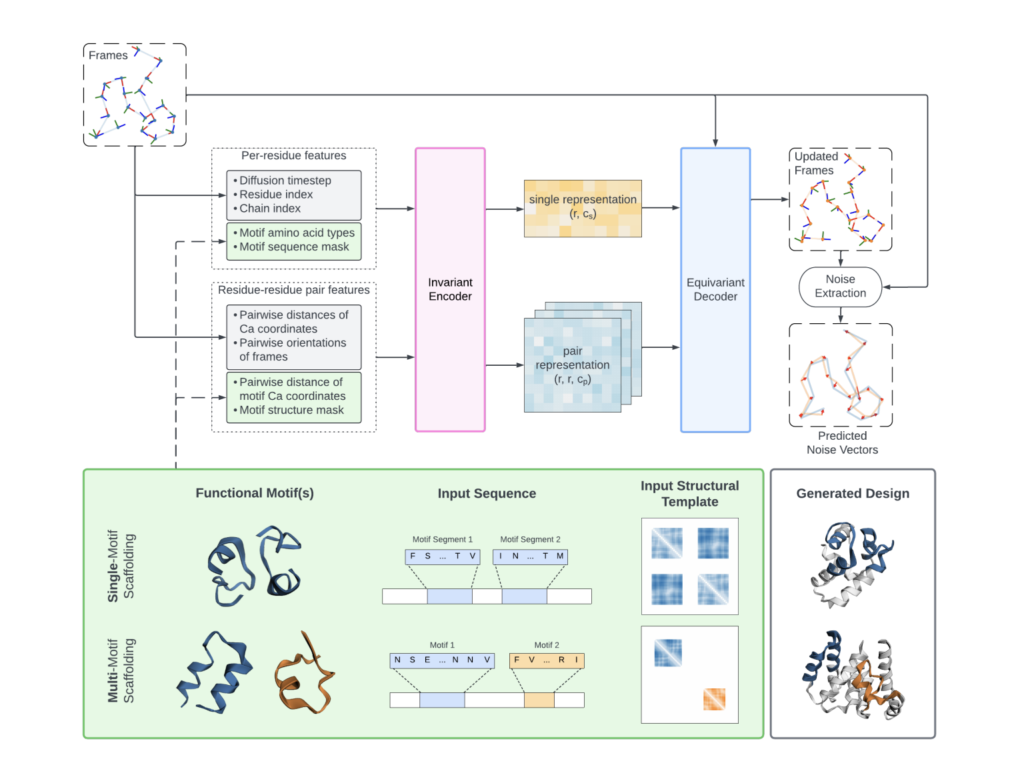
Researchers from Columbia University and Rutgers University introduced Genie 2, an advanced protein design model that extends the capabilities of its predecessor, Genie. Developed by Columbia University and Rutgers University, Genie 2 incorporates architectural innovations and data augmentation to capture a broader protein structure space and enables multi-motif scaffolding for complex protein designs. This new model represents proteins as point clouds of C-alpha atoms in the forward process and clouds of reference frames in the reverse process, enhancing its ability to design complex protein structures.
Genie 2 utilizes SE(3)-equivariant attention mechanisms and asymmetric protein representations in its forward and reverse diffusion processes. It encodes motifs using pairwise distance matrices and integrates these into the diffusion model, allowing the generation of proteins with multiple, independent functional sites without predefined inter-motif positions. This approach sidesteps challenges in multi-motif scaffolding, enabling the design of proteins with complex interaction patterns and multiple functional motifs. The training process involves data augmentation using a subset of the AlphaFold database, consisting of approximately 214 million predictions, significantly enhancing the model’s capabilities.
Genie 2’s performance achieves state-of-the-art designability, diversity, and novelty results. It outperforms existing models like RFDiffusion and FrameFlow in unconditional protein generation and motif scaffolding tasks. For example, Genie 2 achieves a designability score of 0.96, compared to RFDiffusion’s 0.63, and exhibits higher structural diversity and novelty. The model also solves motif scaffolding problems with unique and varied solutions, demonstrating its superior ability to generate complex protein designs.

In conclusion, Genie 2 addresses significant challenges in protein design by introducing a robust model capable of generating complex, multifunctional proteins. It sets a new standard in the field, offering promising tools for future applications in biotechnology and medicine. The researchers’ advancements in architectural innovations and data augmentation techniques have resulted in a model that achieves high performance and broadens the potential for designing novel proteins with specific functional properties.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 43k+ ML SubReddit | Also, check out our AI Events Platform
![]()
Aswin AK is a consulting intern at MarkTechPost. He is pursuing his Dual Degree at the Indian Institute of Technology, Kharagpur. He is passionate about data science and machine learning, bringing a strong academic background and hands-on experience in solving real-life cross-domain challenges.